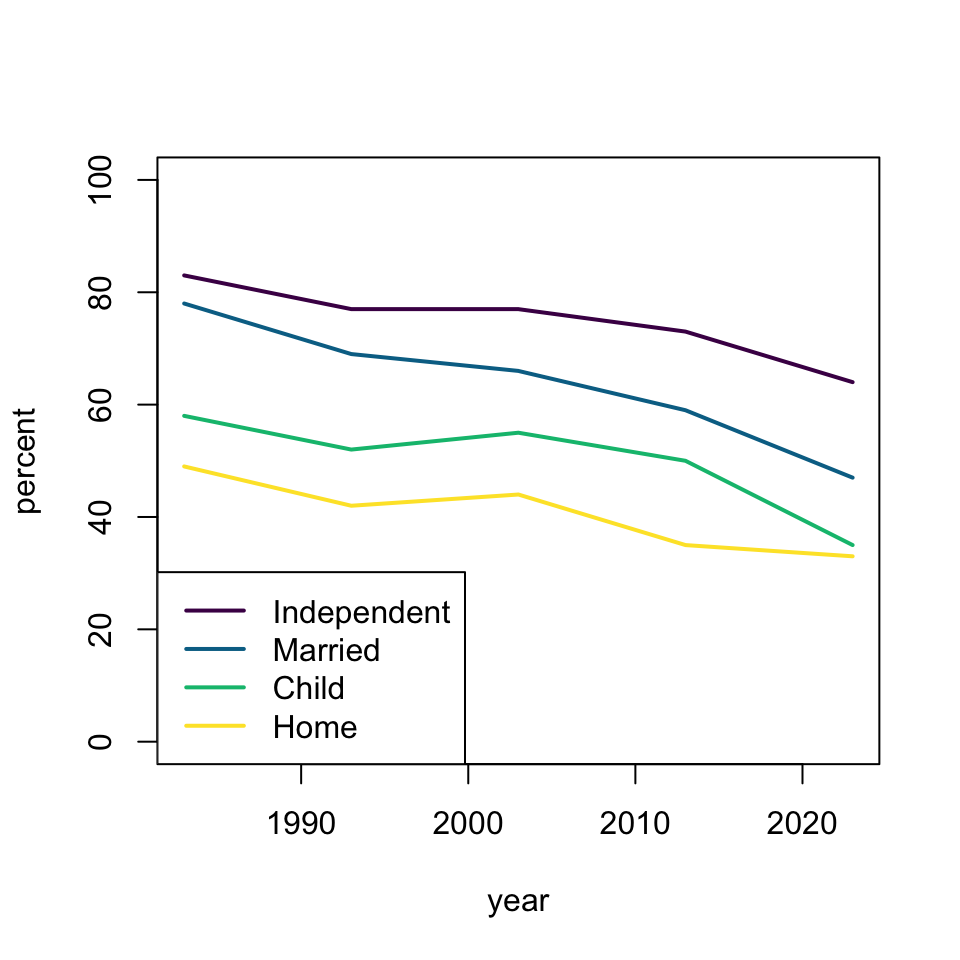
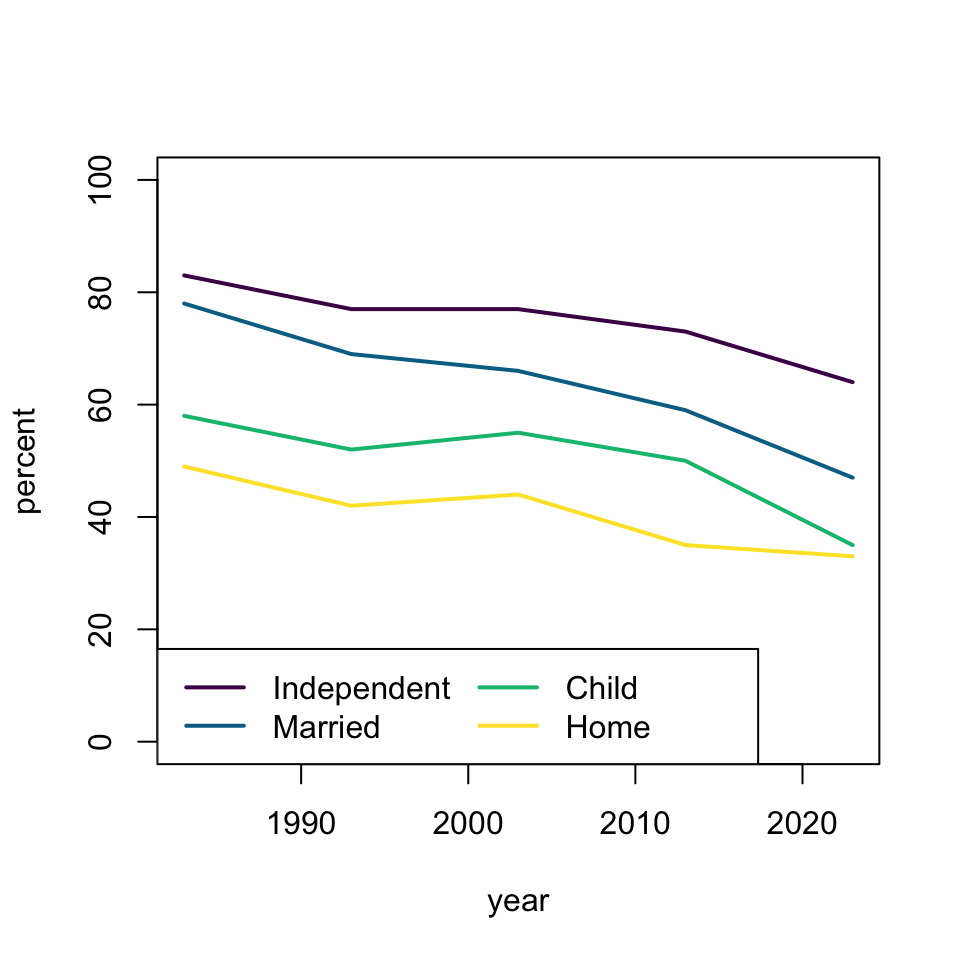
plot(percent ~ year, dat, subset = milestone == "independent", ylim = c(0, 100), type = 'l', lwd = 2, col = hcl.colors(4)[1])
lines(percent ~ year, dat, subset = milestone == "married", type = 'l', lwd = 2, col = hcl.colors(4)[2])
lines(percent ~ year, dat, subset = milestone == "child", type = 'l', lwd = 2, col = hcl.colors(4)[3])
lines(percent ~ year, dat, subset = milestone == "home", type = 'l', lwd = 2, col = hcl.colors(4)[4])
legend("bottomleft", c("Independent", "Married", "Child", "Home"), lty = 1, lwd = 2, col = hcl.colors(4))